Interface with ASE & pymatgen¶
Supported versions of ASE/pymatgen can be installed from the requirements/optional-requirements.txt file, or via pip install matador-db[optional]. This allows for matador.crystal.Crystal objects to be converted to ase.Atoms and pymatgen.Structure objects, as shall be showcased below.
First, let’s use the builder functionality from ASE to create a crystal structure (example from the ase.build docs).
[1]:
import ase.build
gnr1 = ase.build.graphene_nanoribbon(
3, 4, type='armchair', saturated=True, vacuum=3.5
)
print(gnr1)
Atoms(symbols='C48H16', pbc=[False, False, True], cell=[15.03671574711959, 7.0, 17.04])
Let’s attach some metadata using the Atoms.info property:
[2]:
gnr1.info = {
"source": "ase.build.graphene_nanoribbon",
"chiral_vector": (3, 4),
"saturated": True,
"vacuum": 3.5
}
We can now convert this to a matador-style dictionary, or a matador.crystal.Crystal object:
[3]:
from matador.utils.ase_utils import ase2dict
from matador.crystal import Crystal
gnr1_doc = ase2dict(gnr1)
print(gnr1_doc.keys())
print('----')
gnr1_crystal = ase2dict(gnr1, as_model=True)
print(gnr1_crystal)
dict_keys(['atom_types', 'positions_frac', 'lattice_cart', 'lattice_abc', 'num_atoms', 'stoichiometry', 'cell_volume', 'elems', 'num_fu', 'space_group', 'ase_info'])
----
C₃H: unknown
============
64 atoms. Pmma
(a, b, c) = 15.0367 Å, 7.0000 Å, 17.0400 Å
(α, β, γ) = 90.0000° 90.0000° 90.0000°
In the Crystal print out, the “unknown” means that matador could not find any metadata regarding the source of the structure, so let’s use the ASE metadata to update that.
[4]:
# all underlying data will be copied to the `_data` attribute of the Crystal,
# if we want, these keys can be promoted to the actual Crystal data itself
gnr1_crystal.update(gnr1_crystal._data["ase_info"])
print(gnr1_crystal)
C₃H: ase.build.graphene_nanoribbon
==================================
64 atoms. Pmma
(a, b, c) = 15.0367 Å, 7.0000 Å, 17.0400 Å
(α, β, γ) = 90.0000° 90.0000° 90.0000°
[5]:
print(gnr1_crystal.pdf.spg)
Pmma
We can now use matador functionality on this graphene nanoribbon:
[6]:
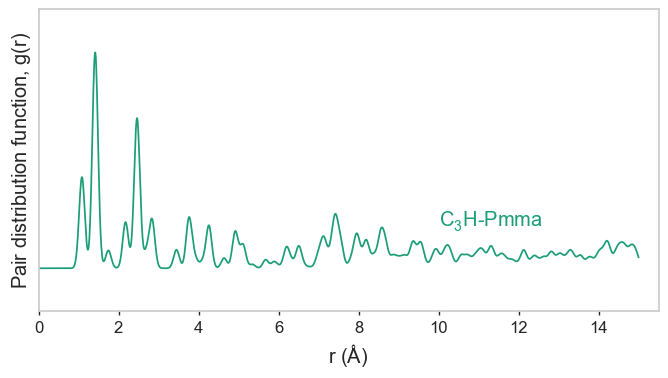
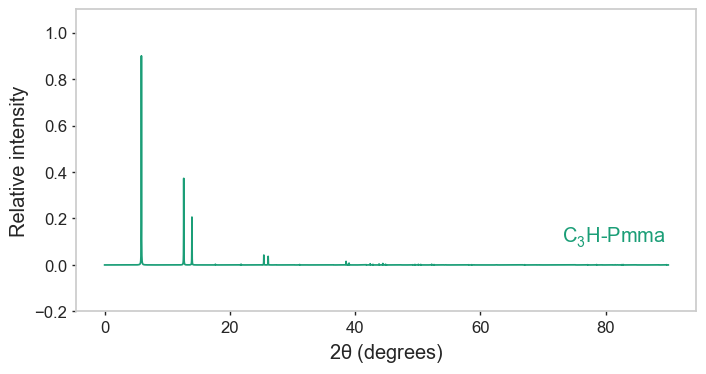
gnr1_crystal.pdf.plot(text_offset=(10, 0.2))
gnr1_crystal.pxrd.plot()
[7]:
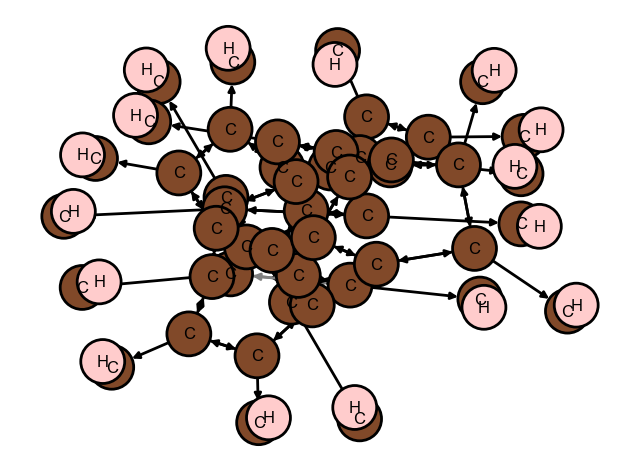
from matador.crystal.network import draw_network
draw_network(gnr1_crystal.network)
print(gnr1_crystal.bonding_stats[0])
Calculated: 110592, Used: 1616, Ignored: 108976
Calculated distances in 0.003461122512817383 s
{'species': 'C', 'position': array([0.62267533, 0.5 , 0. ]), 'bonds': [{'species': 'C', 'index': 1, 'length': 1.4199999999999997, 'is_image': False, 'position': array([0.70445889, 0.5 , 0.04166667])}, {'species': 'C', 'index': 15, 'length': 1.42, 'is_image': True, 'position': array([0.62267533, 0.5 , 0.91666667])}, {'species': 'C', 'index': 17, 'length': 1.4199999999999997, 'is_image': False, 'position': array([0.54089178, 0.5 , 0.04166667])}]}
We can now convert this Crystal object freely between ASE and pymatgen. Let’s make a pmg structure from the Crystal, modify it, then make a new Crystal:
[8]:
pmg_structure = gnr1_crystal.pmg_structure
for ind, site in enumerate(pmg_structure):
if ind % 2 == 0:
pmg_structure[ind] = "N"
print(pmg_structure)
Full Formula (H8 C24 N32)
Reduced Formula: HC3N4
abc : 15.036716 7.000000 17.040000
angles: 90.000000 90.000000 90.000000
Sites (64)
# SP a b c
--- ---- -------- --- --------
0 N 0.622675 0.5 0
1 C 0.704459 0.5 0.041667
2 N 0.704459 0.5 0.125
3 C 0.622675 0.5 0.166667
4 N 0.622675 0.5 0.25
5 C 0.704459 0.5 0.291667
6 N 0.704459 0.5 0.375
7 C 0.622675 0.5 0.416667
8 N 0.622675 0.5 0.5
9 C 0.704459 0.5 0.541667
10 N 0.704459 0.5 0.625
11 C 0.622675 0.5 0.666667
12 N 0.622675 0.5 0.75
13 C 0.704459 0.5 0.791667
14 N 0.704459 0.5 0.875
15 C 0.622675 0.5 0.916667
16 N 0.459108 0.5 0
17 C 0.540892 0.5 0.041667
18 N 0.540892 0.5 0.125
19 C 0.459108 0.5 0.166667
20 N 0.459108 0.5 0.25
21 C 0.540892 0.5 0.291667
22 N 0.540892 0.5 0.375
23 C 0.459108 0.5 0.416667
24 N 0.459108 0.5 0.5
25 C 0.540892 0.5 0.541667
26 N 0.540892 0.5 0.625
27 C 0.459108 0.5 0.666667
28 N 0.459108 0.5 0.75
29 C 0.540892 0.5 0.791667
30 N 0.540892 0.5 0.875
31 C 0.459108 0.5 0.916667
32 N 0.295541 0.5 0
33 C 0.377325 0.5 0.041667
34 N 0.377325 0.5 0.125
35 C 0.295541 0.5 0.166667
36 N 0.295541 0.5 0.25
37 C 0.377325 0.5 0.291667
38 N 0.377325 0.5 0.375
39 C 0.295541 0.5 0.416667
40 N 0.295541 0.5 0.5
41 C 0.377325 0.5 0.541667
42 N 0.377325 0.5 0.625
43 C 0.295541 0.5 0.666667
44 N 0.295541 0.5 0.75
45 C 0.377325 0.5 0.791667
46 N 0.377325 0.5 0.875
47 C 0.295541 0.5 0.916667
48 N 0.232764 0.5 0.031984
49 H 0.232764 0.5 0.134683
50 N 0.232764 0.5 0.281984
51 H 0.232764 0.5 0.384683
52 N 0.232764 0.5 0.531984
53 H 0.232764 0.5 0.634683
54 N 0.232764 0.5 0.781984
55 H 0.232764 0.5 0.884683
56 N 0.767236 0.5 0.009683
57 H 0.767236 0.5 0.156984
58 N 0.767236 0.5 0.259683
59 H 0.767236 0.5 0.406984
60 N 0.767236 0.5 0.509683
61 H 0.767236 0.5 0.656984
62 N 0.767236 0.5 0.759683
63 H 0.767236 0.5 0.906984
/home/mevans/.local/conda/envs/matador_thesis/lib/python3.8/site-packages/pymatgen/core/__init__.py:49: UserWarning: Error loading .pmgrc.yaml: [Errno 2] No such file or directory: '/home/mevans/.pmgrc.yaml'. You may need to reconfigure your yaml file.
warnings.warn(f"Error loading .pmgrc.yaml: {ex}. You may need to reconfigure your yaml file.")
[9]:
gnr1_doped_crystal = Crystal.from_pmg(pmg_structure)
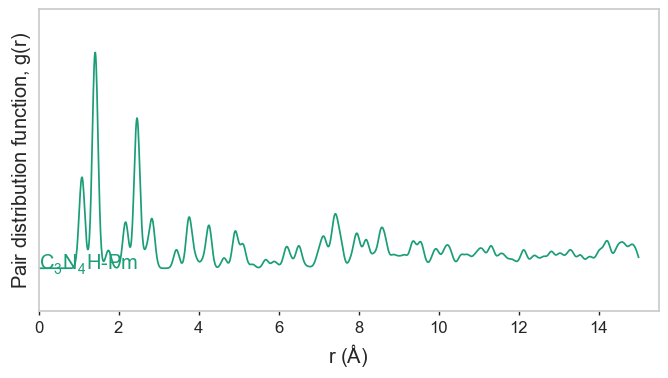
Now we can plot the C-N, N-N and H-N pair distribution functions:
[10]:
gnr1_doped_crystal.pdf.plot_projected_pdf(keys=[("N", ), ("C", "N"), ("H", "N")])
[11]:
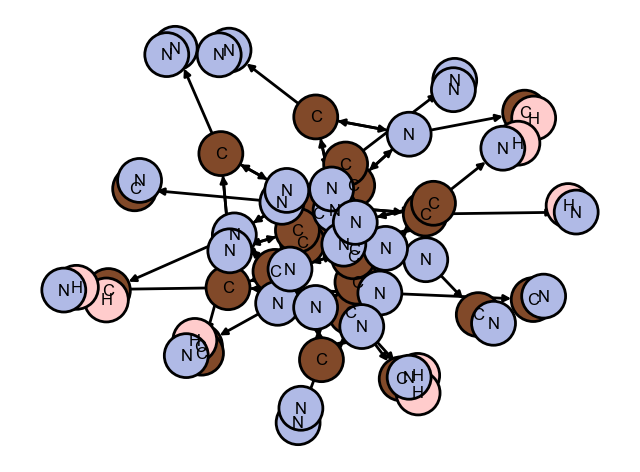
draw_network(gnr1_doped_crystal.network)
Calculated: 110592, Used: 1616, Ignored: 108976
Calculated distances in 0.003873586654663086 s