Analysing and plotting magnetic resonance data¶
This notebook contains a quick example of plotting data stored in magres files, without performing any referencing.
[1]:
from matador.scrapers import magres2dict
from matador.plotting import plot_magres
magres, failures = magres2dict("*.magres", as_model=True)
Successfully scraped 2 out of 2 files.
[2]:
for doc in magres:
print(doc)
for site in doc:
print(site)
break
Li₃P: LiP_CASTEP18
==================
20 atoms. P1
(a, b, c) = 4.1333 Å, 6.0638 Å, 12.5450 Å
(α, β, γ) = 88.4518° 80.5130° 90.0085°
Li 0.0916 0.3584 0.9424
chemical_shielding_iso = 83.7003
magnetic_shielding_tensor =
[[ 8.16e+01 6.11e-03 -4.30e-02]
[ 1.27e-02 7.99e+01 1.36e+00]
[-5.07e-02 2.13e+00 8.96e+01]]
chemical_shift_aniso = 9.3772
chemical_shift_asymmetry = 0.3284
---
Na₃P: NaP_QE6
=============
4 atoms. Fm-3m
(a, b, c) = 5.0075 Å, 5.0075 Å, 5.0075 Å
(α, β, γ) = 120.0020° 120.0020° 89.9965°
Na 0.0000 0.0000 0.0000
chemical_shielding_iso = 518.1528
magnetic_shielding_tensor =
[[518.53 -0. 0. ]
[ -0. 518.53 0. ]
[ -0. 0. 517.4 ]]
chemical_shift_aniso = -1.1365
chemical_shift_asymmetry = -0.0000
---
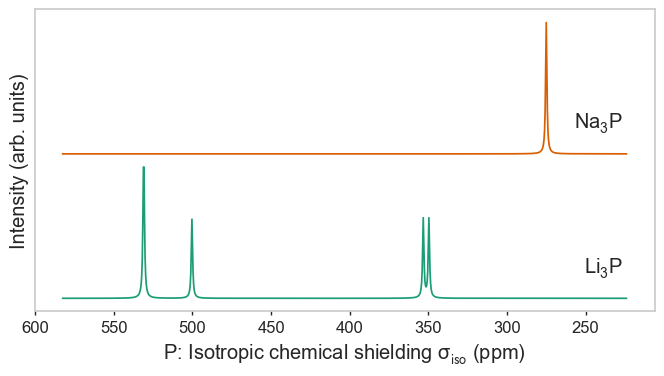
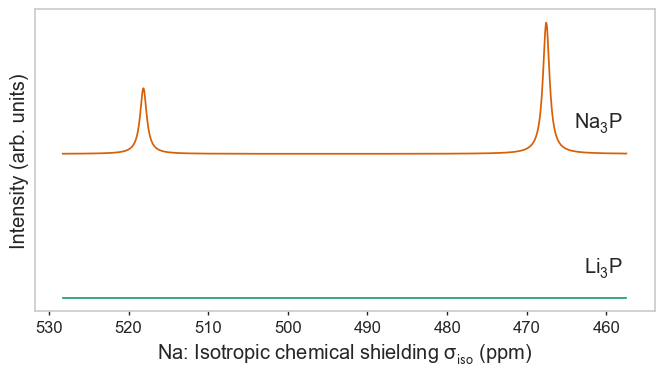
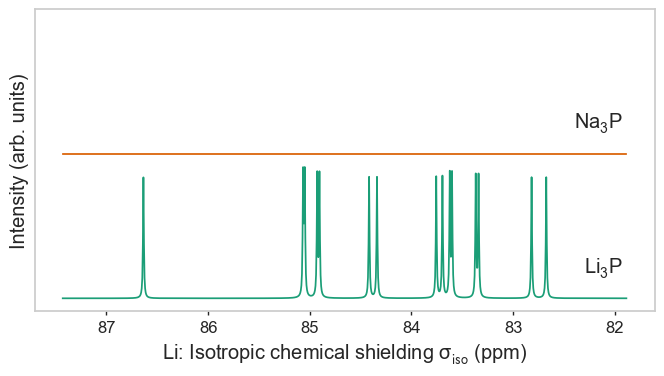
Species as separate figures¶
[3]:
plot_magres(magres, "P")
plot_magres(magres, "Na")
plot_magres(magres, "Li", broadening_width=0.01);
No sites of Na found in LiP_CASTEP18, signal will be empty.
No sites of Li found in NaP_QE6, signal will be empty.
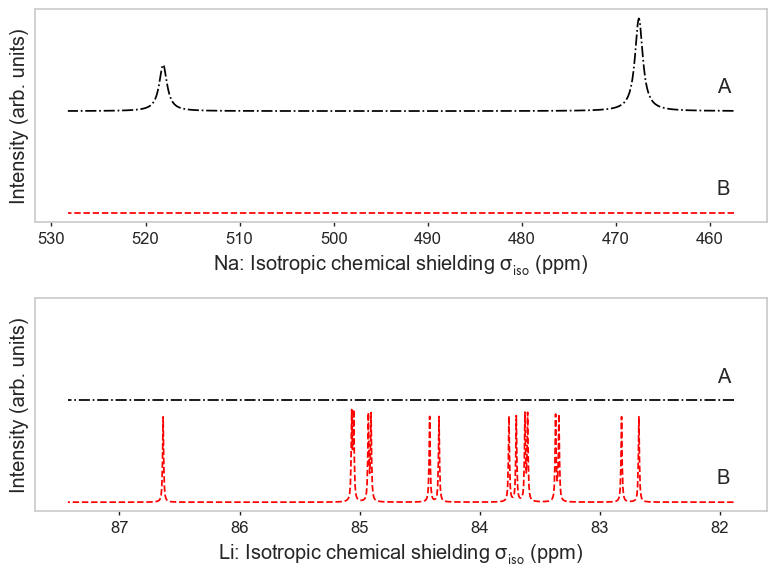
Species as subplots with custom colours and labels¶
[4]:
import matplotlib.pyplot as plt
fig, axes = plt.subplots(2, 1, figsize=(8, 6))
line_kwargs = [{"c": "red", "ls": "--"}, {"c": "black", "ls": "-."}]
labels = ["B", "A"]
plot_magres(
magres, "Na",
ax=axes[0], line_kwargs=line_kwargs, signal_labels=labels
)
plot_magres(
magres, "Li", broadening_width=0.01,
ax=axes[1], line_kwargs=line_kwargs, signal_labels=labels
)
plt.tight_layout()
No sites of Na found in LiP_CASTEP18, signal will be empty.
No sites of Li found in NaP_QE6, signal will be empty.
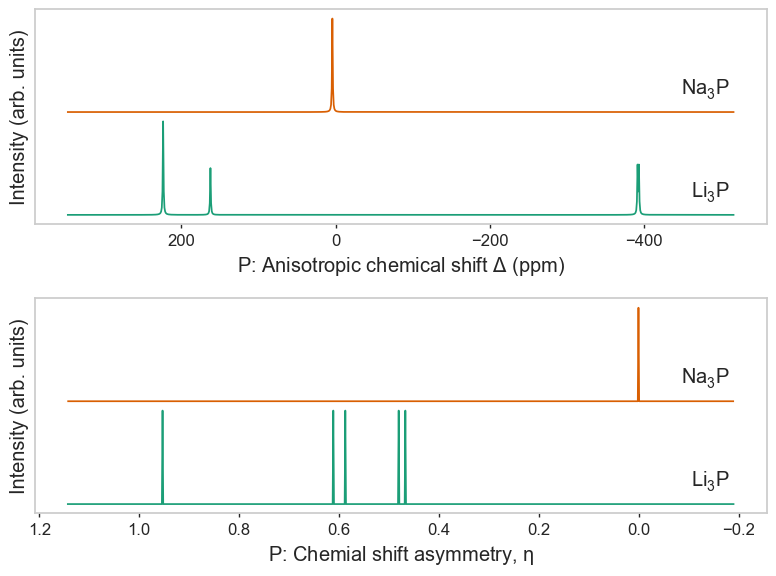
Other magres quantities as subplots¶
[5]:
import matplotlib.pyplot as plt
fig, axes = plt.subplots(2, 1, figsize=(8, 6))
plot_magres(
magres, "P", magres_key="chemical_shift_aniso",
ax=axes[0]
)
plot_magres(
magres, "P", magres_key="chemical_shift_asymmetry",
ax=axes[1],
broadening_width=0,
)
plt.tight_layout()