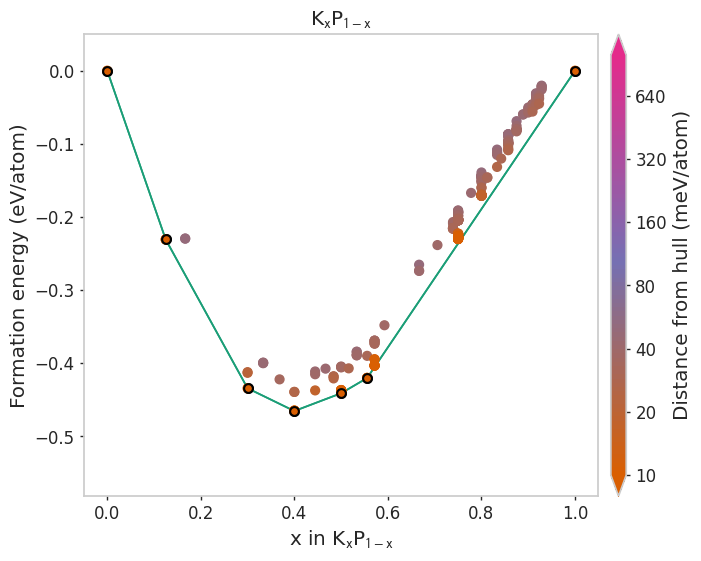
Plotting styles in matador¶
This notebook shows how to alter plotting styles in matador. Typically, these results would be achieved by editing the default_style field in your .matadorrc file. Finer grained control can be obtained using the API (see section on patching styles). Any function that is decorated with @plotting_function will have these styles applied.
[1]:
%matplotlib inline
import glob
import os
import matplotlib.pyplot as plt
[2]:
from matador.plotting import plot_2d_hull
from matador.plotting import plot_spectral
from matador.hull import QueryConvexHull
from matador.scrapers import res2dict
[3]:
cursor, failures = res2dict(glob.glob("hull_data/*.res"))
if cursor is None:
raise RuntimeError("Wrong directory requested, no files found...")
hull = QueryConvexHull(
cursor=cursor,
species="KP",
no_plot=True
)
Successfully scraped 295 out of 295 files.
7 structures found within 0.0 eV of the hull, including chemical potentials.
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Source !?! Pressure Cell volume Hull dist. Space group Formula # fu Prov.
(GPa) (ų/fu) (meV/atom)
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
* K-Na-Collo 0.02 73.3 0.0 Im-3m K 1 SWAPS
* KP-NaP-CollCode56530 -0.00 267.4 0.0 C2/m K₅P₄ 1 SWAPS
* KP-GA-scno1g-3x2 0.05 57.4 0.0 P1 KP 8 GA
* KP-GA-0ss21w-17x39 0.01 147.8 0.0 P1 K₂P₃ 4 GA
* KP-Cs3P7-OQMD_57401-CollCode62259 -0.00 294.8 0.0 P4_1 K₃P₇ 4 SWAPS
* KP-LiP-ColCode23621 0.01 229.8 0.0 I4_1/acd KP₇ 8 SWAPS
* P-CollCode150873 -0.01 21.2 0.0 Cmce P 4 ICSD
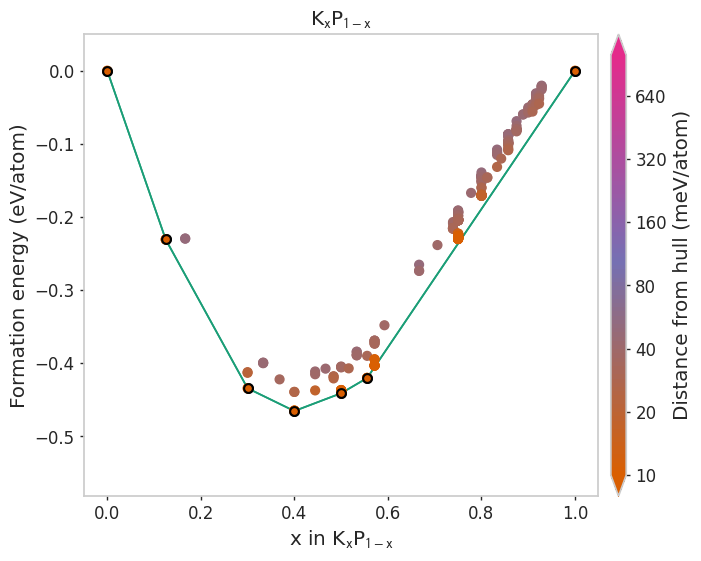
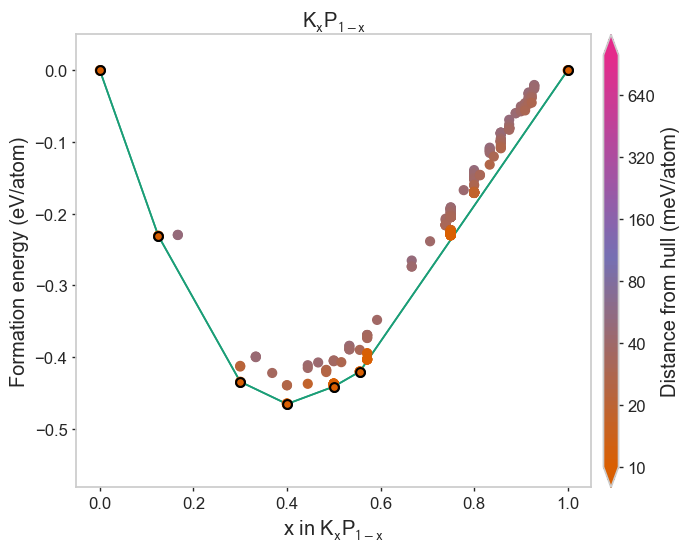
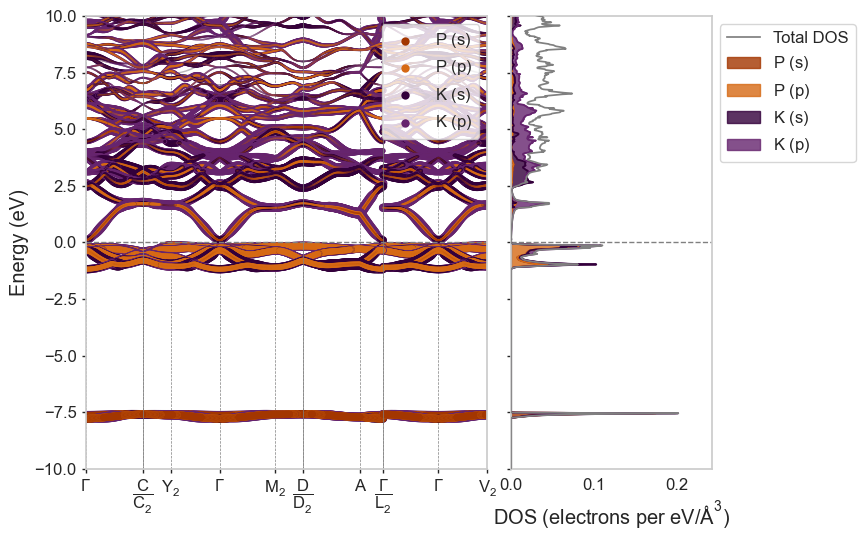
Using different built-in styles¶
[4]:
styles = ['matador', 'seaborn', 'dark_background', 'default']
for style in styles:
# reset to default to avoid stacking plot styles
if hasattr(hull, 'colours'):
del hull.colours
plot_2d_hull(hull, style=style)
plot_spectral(
"dispersion_data/K3P-OQMD_4786-CollCode25550",
style=style
)
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1061: UserWarning: Warning: converting a masked element to nan.
flat_zorders = np.asarray(flat_zorders)
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1064: UserWarning: Warning: converting a masked element to nan.
flat_sizes = np.asarray(flat_sizes)[np.argsort(flat_zorders)]
Unable to create spglib structure from input data: skipping path labels: Unable to use doc2spg, one of ('lattice_cart', 'positions_frac', 'atom_types') was missing..
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:329: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout()
Displaying plot...
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1061: UserWarning: Warning: converting a masked element to nan.
flat_zorders = np.asarray(flat_zorders)
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1064: UserWarning: Warning: converting a masked element to nan.
flat_sizes = np.asarray(flat_sizes)[np.argsort(flat_zorders)]
Unable to create spglib structure from input data: skipping path labels: Unable to use doc2spg, one of ('lattice_cart', 'positions_frac', 'atom_types') was missing..
Displaying plot...
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:329: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout()
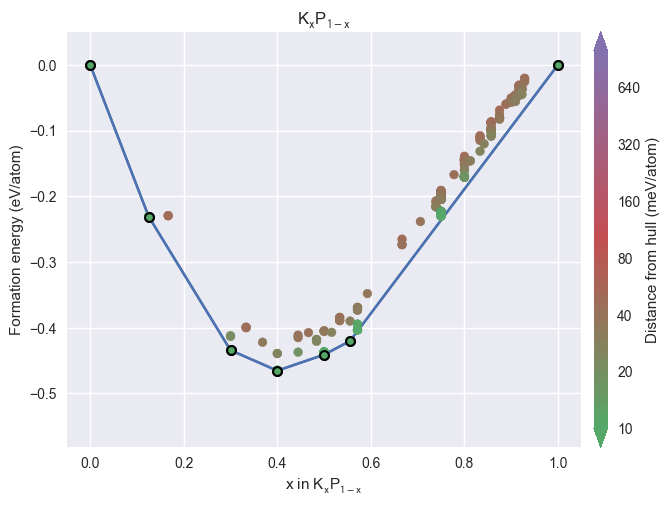
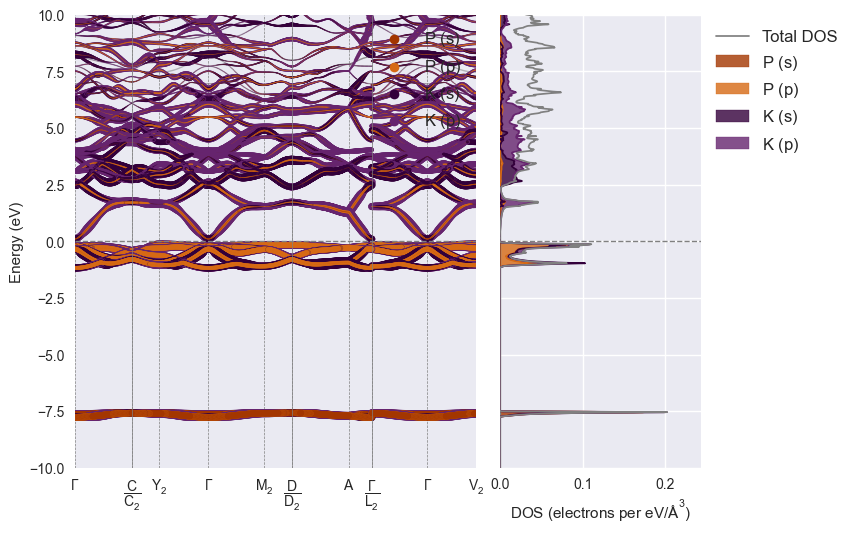
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1061: UserWarning: Warning: converting a masked element to nan.
flat_zorders = np.asarray(flat_zorders)
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1064: UserWarning: Warning: converting a masked element to nan.
flat_sizes = np.asarray(flat_sizes)[np.argsort(flat_zorders)]
Unable to create spglib structure from input data: skipping path labels: Unable to use doc2spg, one of ('lattice_cart', 'positions_frac', 'atom_types') was missing..
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:329: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout()
Displaying plot...
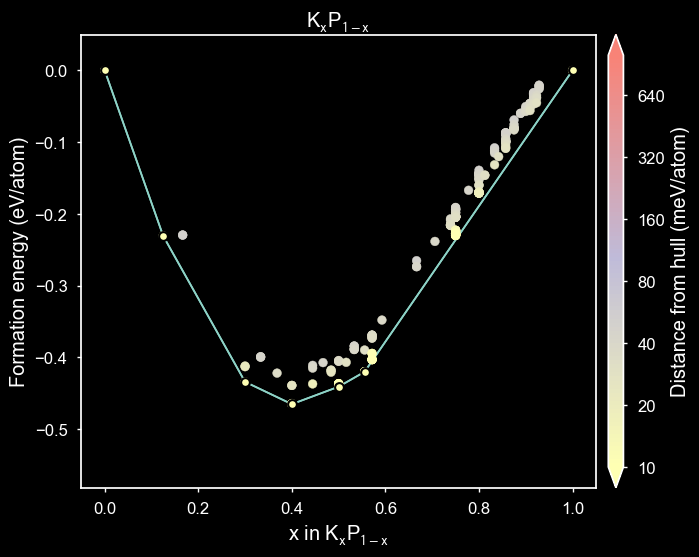
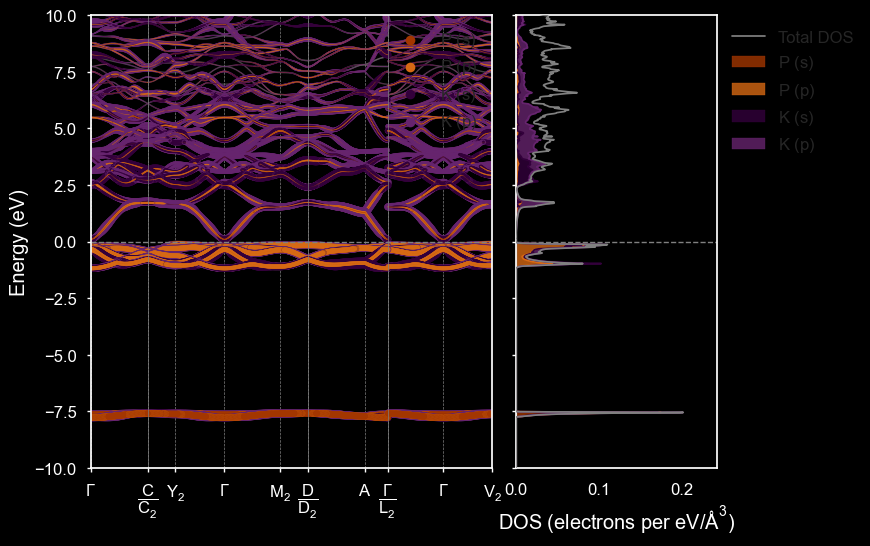
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1061: UserWarning: Warning: converting a masked element to nan.
flat_zorders = np.asarray(flat_zorders)
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:1064: UserWarning: Warning: converting a masked element to nan.
flat_sizes = np.asarray(flat_sizes)[np.argsort(flat_zorders)]
Unable to create spglib structure from input data: skipping path labels: Unable to use doc2spg, one of ('lattice_cart', 'positions_frac', 'atom_types') was missing..
Displaying plot...
/home/mevans/src/matador/matador/plotting/spectral_plotting.py:329: UserWarning: This figure includes Axes that are not compatible with tight_layout, so results might be incorrect.
plt.tight_layout()
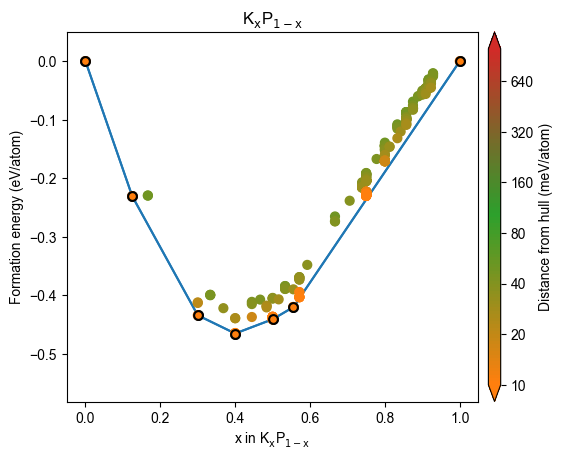
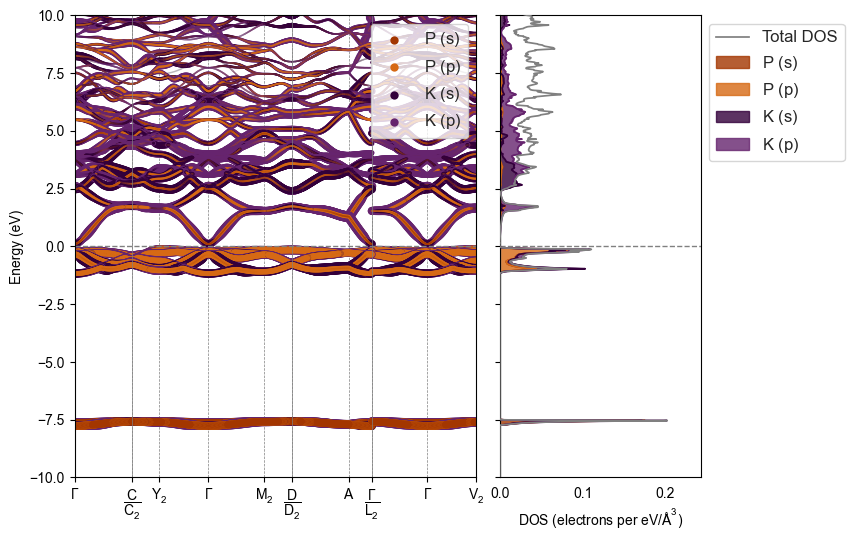
Patching styles on demand¶
[5]:
fonts = ['Arial Black', 'Archivo Narrow']
for font in fonts:
# reset to default to avoid stacking plot styles
if hasattr(hull, 'colours'):
del hull.colours
plot_2d_hull(hull, style=['matador', {'font.sans-serif': [font]}])
findfont: Font family ['sans-serif'] not found. Falling back to DejaVu Sans.
findfont: Generic family 'sans-serif' not found because none of the following families were found: Archivo Narrow
findfont: Font family ['sans-serif'] not found. Falling back to DejaVu Sans.
findfont: Generic family 'sans-serif' not found because none of the following families were found: Archivo Narrow