Creating a convex hull and extracting some data¶
This example takes data from our private group database, creates a compositional phase diagram, then extracts it into a matador-agnostic format.
[1]:
%matplotlib inline
from matador.query import DBQuery
from matador.hull import QueryConvexHull
[2]:
query = DBQuery(
composition='NaP',
summary=True,
subcmd='hull',
)
hull = QueryConvexHull(
query,
no_plot=False,
hull_cutoff=26e-3,
summary=True,
labels=True,
)
Loading settings from /home/mevans/.matadorrc
2324 results found for query in repo.
Creating hull from structures in query results.
Finding the best calculation set for hull...
component veil : matched 1253 structures. -> PBE 800.0 eV, xxx eV/A, 0.03 1/A.
siege toys : matched 1070 structures. -> PBE 400.0 eV, 0.05 eV/A, 0.04 1/A.
comprise toad : matched 1070 structures. -> PBE 400.0 eV, 0.05 eV/A, 0.05 1/A.
furlough road : matched 1253 structures. -> PBE 800.0 eV, xxx eV/A, 0.03 1/A.
sanctity toys : matched 1253 structures. -> PBE 800.0 eV, xxx eV/A, 0.03 1/A.
Composing hull from set containing component veil
────────────────────────────────────────────────────────────
Scanning for suitable Na chemical potential...
Using irreducible tomatoes as chem pot for Na
────────────────────────────────────────────────────────────
Scanning for suitable P chemical potential...
Using personage daughter as chem pot for P
────────────────────────────────────────────────────────────
87 structures found within 0.026 eV of the hull, including chemical potentials.
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
ID !?! Pressure Cell volume Hull dist. Space group Formula # fu Prov. Occurrences
(GPa) (ų/fu) (meV/atom)
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
* irreducible tomatoes 0.00 37.2 0.0 Im-3m Na 1 ICSD
* precession screw 0.01 94.9 0.0 P63cm Na3P 6 ICSD
disinfect pencil -0.07 254.6 25.8 Cm Na7P4 1 GA
incidentally men -0.01 155.6 1.6 Cmcm Na4P3 2 ICSD
* dissonance part 0.00 197.5 0.0 C2/m Na5P4 1 ICSD
* corrosive experience 0.02 42.3 0.0 P21/c NaP 8 ICSD
virtu camp -0.02 65.0 20.1 C2/m NaP2 2 ICSD
* intermittent pan 0.00 253.7 0.0 P212121 Na3P7 4 ICSD
ravine shoes -0.04 1009.3 25.0 P1 Na11P28 1 GA
* soliloquy kick -0.03 329.7 0.0 Pbcn Na3P11 4 ICSD
embellish furniture -0.00 128.0 17.0 Pnma NaP5 4 ICSD
* flagrant crook 0.01 194.0 0.0 I41/acd NaP7 8 AIRSS
* personage daughter 0.01 21.1 0.0 Cmca P 4 ICSD
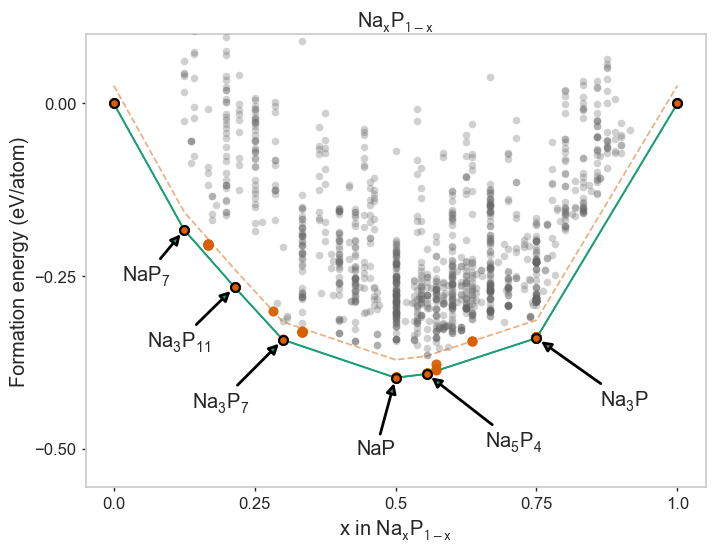
[3]:
hull.voltage_curve()
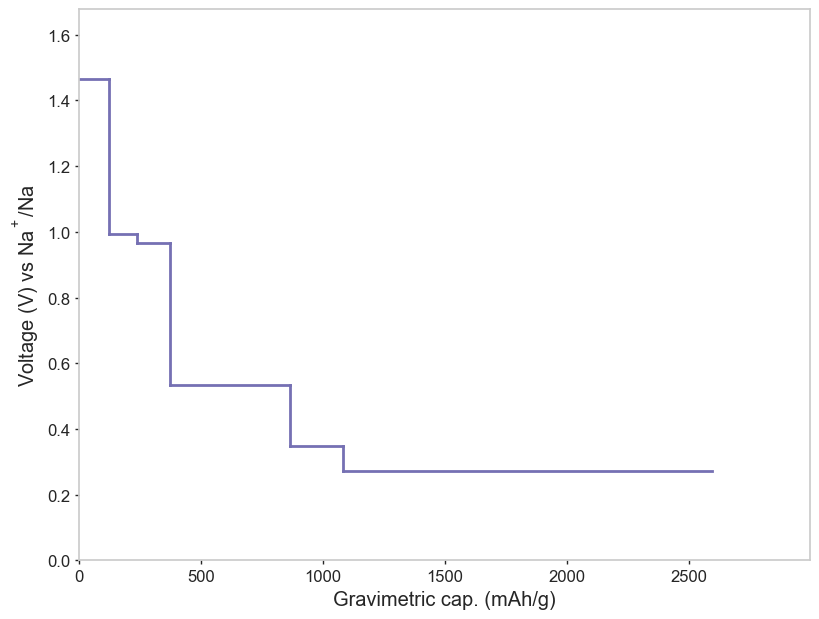
hull.plot_voltage_curve(labels=False)
Voltage data:
# Na into P
# Average voltage: 0.45 V
# Q (mAh/g) Voltage (V)
0.00 1.46589125
123.62 1.46589125
236.01 0.99442375
370.87 0.96562208
865.37 0.53491063
1081.71 0.34735000
2596.10 0.27326571
nan 0.00000000
[4]:
# see what keys are available in each structure
print(hull.hull_cursor[0].keys())
dict_keys(['_id', 'task', 'total_energy_per_atom', 'spin_polarized', 'cut_off_energy', 'enthalpy', 'species_pot', 'geom_method', 'tags', 'space_group', 'metals_method', 'total_energy', 'elems', 'source', 'kpoints_mp_spacing', 'text_id', 'icsd', 'stoichiometry', 'lattice_cart', 'lattice_abc', 'user', 'quality', 'num_fu', 'geom_stress_tol', 'atom_types', 'pressure', 'cell_volume', 'num_atoms', 'enthalpy_per_atom', 'external_pressure', 'positions_frac', 'xc_functional', 'ratios', 'enthalpy_per_b', 'num_a', 'num_chempots', 'concentration', 'formation_enthalpy_per_atom', 'formation_enthalpy', 'hull_distance', 'cell_volume_per_b', 'gravimetric_capacity'])
[5]:
from matador.utils.cursor_utils import get_array_from_cursor
# here we grab the first element of the concentration array, which only contains 1 element for a binary system
concentrations = get_array_from_cursor(hull.hull_cursor, ['concentration', 0])
formation_enthalpy = get_array_from_cursor(hull.hull_cursor, 'formation_enthalpy_per_atom')
hull_distances = get_array_from_cursor(hull.hull_cursor, 'hull_distance')
for conc, eform, hdist in zip(concentrations, formation_enthalpy, hull_distances):
print(f"{conc:12.4f} {eform:12.4f} {1000*hdist:12.4f}")
0.0000 0.0000 0.0000
0.1250 -0.1832 0.0000
0.1667 -0.2049 16.9654
0.1667 -0.2048 17.0896
0.1667 -0.2047 17.1937
0.1667 -0.2040 17.8146
0.1667 -0.2040 17.8225
0.1667 -0.2040 17.8500
0.2143 -0.2660 0.0000
0.2143 -0.2655 0.5179
0.2821 -0.3013 25.0358
0.3000 -0.3423 0.0000
0.3000 -0.3423 0.0700
0.3000 -0.3422 0.0900
0.3000 -0.3420 0.3225
0.3000 -0.3419 0.4525
0.3333 -0.3314 20.1031
0.3333 -0.3311 20.3548
0.3333 -0.3311 20.4081
0.3333 -0.3306 20.9148
0.3333 -0.3301 21.4181
0.5000 -0.3974 0.0000
0.5000 -0.3973 0.0125
0.5000 -0.3973 0.0187
0.5000 -0.3973 0.0438
0.5000 -0.3973 0.0625
0.5000 -0.3973 0.0625
0.5000 -0.3973 0.0750
0.5000 -0.3972 0.1062
0.5000 -0.3972 0.1125
0.5000 -0.3970 0.3937
0.5000 -0.3967 0.6750
0.5000 -0.3967 0.6875
0.5000 -0.3966 0.7625
0.5000 -0.3966 0.7688
0.5556 -0.3918 0.0000
0.5556 -0.3918 0.0433
0.5556 -0.3912 0.6211
0.5556 -0.3912 0.6422
0.5556 -0.3911 0.6778
0.5556 -0.3910 0.7678
0.5714 -0.3860 1.5959
0.5714 -0.3853 2.2173
0.5714 -0.3772 10.3923
0.6364 -0.3444 25.8210
0.6364 -0.3443 25.9247
0.7500 -0.3399 0.0000
0.7500 -0.3396 0.2958
0.7500 -0.3396 0.3000
0.7500 -0.3396 0.3375
0.7500 -0.3396 0.3417
0.7500 -0.3394 0.5458
0.7500 -0.3393 0.6042
0.7500 -0.3392 0.7708
0.7500 -0.3391 0.8625
0.7500 -0.3388 1.1750
0.7500 -0.3387 1.2213
0.7500 -0.3387 1.2213
0.7500 -0.3387 1.2225
0.7500 -0.3387 1.2275
0.7500 -0.3387 1.2287
0.7500 -0.3387 1.2287
0.7500 -0.3387 1.2287
0.7500 -0.3387 1.2287
0.7500 -0.3387 1.2287
0.7500 -0.3387 1.2300
0.7500 -0.3387 1.2313
0.7500 -0.3387 1.2337
0.7500 -0.3387 1.2337
0.7500 -0.3387 1.2350
0.7500 -0.3387 1.2375
0.7500 -0.3387 1.2388
0.7500 -0.3387 1.2400
0.7500 -0.3387 1.2413
0.7500 -0.3387 1.2450
0.7500 -0.3387 1.2475
0.7500 -0.3387 1.2500
0.7500 -0.3387 1.2612
0.7500 -0.3387 1.2625
0.7500 -0.3387 1.2700
0.7500 -0.3387 1.2725
0.7500 -0.3387 1.2737
0.7500 -0.3386 1.3062
0.7500 -0.3386 1.3125
0.7500 -0.3384 1.5500
0.7500 -0.3383 1.6562
1.0000 0.0000 0.0000